
"PEPPER: Cytoscape app for Protein complex Expansion using Protein-Protein intERaction networks", *Bioinformatics*, August 2014. This workflow orchestrates **external data integration** (GO annotations, known complexes matching) besides measuring **connectivity features** (topological coefficients). PEPPER further refines predictions by an integrated **structure- and function-based post-processing** pipeline to rank proteins that were added by the algorithm. Supports: Conversion from NetworkX see example1, example2 Conversion from Pandas DataFrame see example Conversion from neo4j see example Installation.
Tooltip cytoscape install#
Try it out using binder: or install and try out the examples. oCytoscape can export its Styles into CSS-based Cytoscape.js JSON. This is a collection of mappings from data point to network property. When importing your own files, make sure that they were saved using UTF-8 encoding. A widget enabling interactive graph visualization with cytoscape.js in JupyterLab and the Jupyter notebook. Cytoscape and Cytoscape.js are sharing a concept called Style. This has been resolved by setting dot-app to import and export files using UTF-8 encoding. Fixes a bug that caused import issues based on characters used in files.

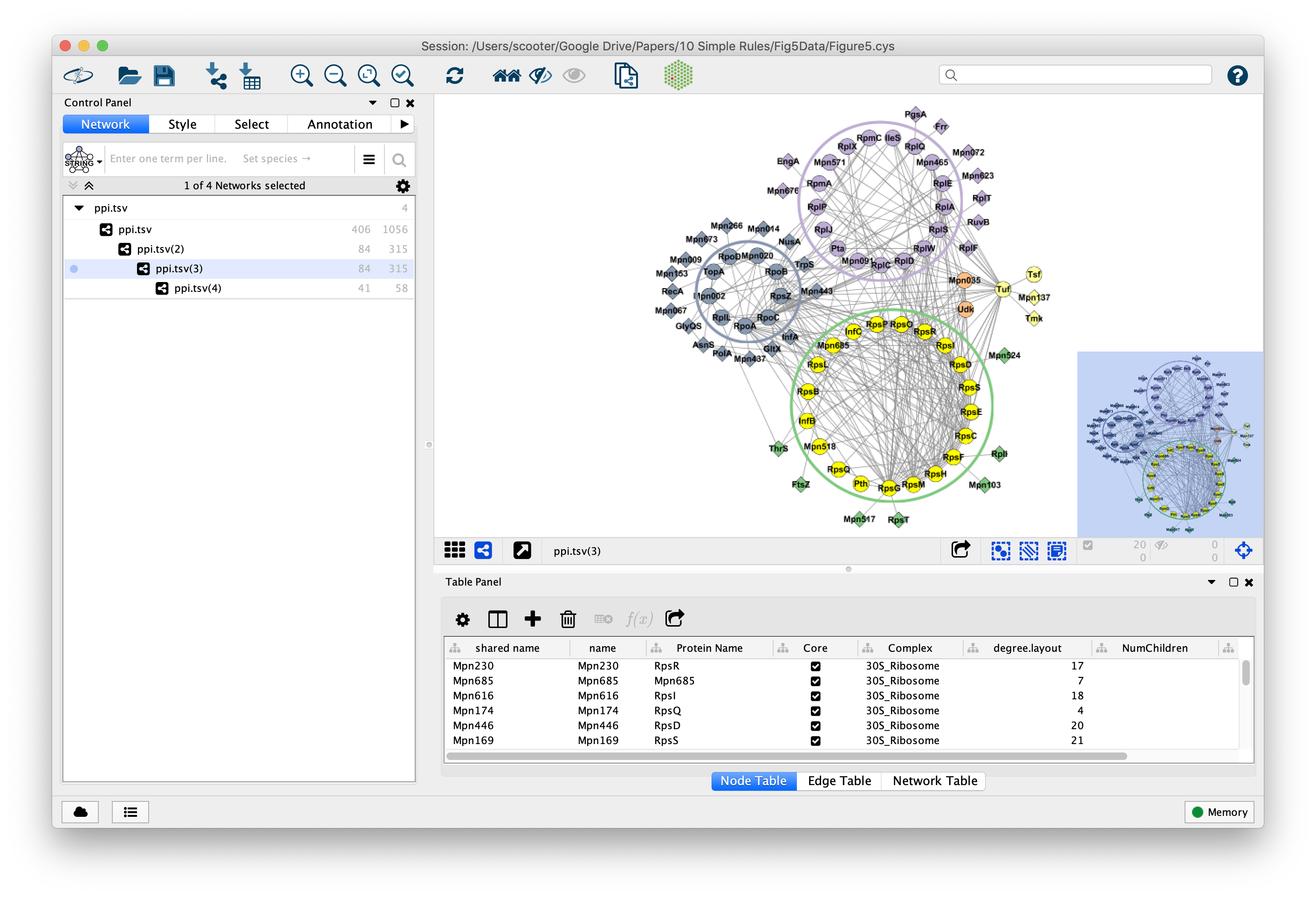
To summarise the information from all solutions, PEPPER merges *Pareto solutions* into a final predicted protein complex by **maximising the modularity** using a greedy search. Fixes a bug that caused tooltips to not be imported. Since these objectives conflict, no single solution can be considered as dominating the others. PEPPER resolves connection sub-graph discovery problems by using **multi-objective optimisation** involving two objective functions: (i) the **coverage**, a solution must contain as many proteins from the *seed* as possible, (ii) the **density**, a solution must contain as many interactions as possible. PEPPER - Protein complex Expansion using Protein-Protein intERaction networks - **identifies meaningful pathways / complexes** as densely connected sub-networks from *seed* lists of proteins derived from pull-down assays (*i.e* AP-MS.).


 0 kommentar(er)
0 kommentar(er)
